Dagstuhl-Seminar 19241
25 Years of the Burrows-Wheeler Transform
( 10. Jun – 14. Jun, 2019 )
Permalink
Organisatoren
- Travis Gagie (Universidad Diego Portales, CL)
- Giovanni Manzini (University of Eastern Piedmont - Alessandria, IT)
- Gonzalo Navarro (University of Chile - Santiago de Chile, CL)
- Jens Stoye (Universität Bielefeld, DE)
Kontakt
- Andreas Dolzmann (für wissenschaftliche Fragen)
- Susanne Bach-Bernhard (für administrative Fragen)
Impacts
- Efficient Construction of a Complete Index for Pan-Genomics Read Alignment : article - Kuhnle, Alan; Mun, Taher; Boucher, Christina; Gagie, Travis; Langmead, Christopher J.; Manzini, Giovanni - New Rochelle, NY : Mary Ann Liebert, Inc., 2019. - pp. 158-173 - (Journal of computational biology ; 27. 2020, 4).
- Fully Functional Suffix Trees and Optimal Text Searching in BWT-Runs Bounded Space : article - Gagie, Travis; Navarro, Gonzalo; Prezza, Nicola - New York : ACM, 2020. - 54 pp. - (Journal of the ACM ; 67. 2020, 2).
- Matching Reads to Many Genomes with the r-Index : article - Mun, Taher; Kuhnle, Alan; Boucher, Christina; Gagie, Travis; Langmead, Christopher J.; Manzini, Giovanni - New Rochelle, NY : Mary Ann Liebert, Inc., 2019. - pp. 158-173 - (Journal of computational biology ; 27. 2020, 4).
- On the Approximation Ratio of Ordered Parsings: article - Navarro, Gonzalo; Ochoa, Carlos; Prezza, Nicola - Cornell University : arXiv.org, 2019. - 21 pp..
- Optimal-Time Dictionary-Compressed Indexes : article - Christiansen, Anders Roy; Ettienne, Mikko Berggren; Kociumaka, Tomasz; Navarro, Gonzalo; Prezza, Nicola - Cornell University : arXiv.org, 2019. - 40 pp. - (ACM Transactions on Algorithms ; Preprint).
- Regular Languages meet Prefix Sorting : article in Proceedings of the 2020 ACM-SIAM Symposium on Discrete Algorithms - Alanko, Jarno; Agostino, Giovanna d`; Policriti, Alberto; Prezza, Nicola - Philadelphia : SIAM, 2020. - 20 pp..
- Text Indexing and Searching in Sublinear Time : article in CPM 2020 - Munro, J. Ian; Navarro, Gonzalo; Nekrich, Yakov - Wadern : LZI, 2020. - pp. 1-15 - (Leibniz International Proceedings in Informatics ; 161 : article)7.
- Towards a Definitive Measure of Repetitiveness : article - Kociumaka, Tomasz; Navarro, Gonzalo; Prezza, Nicola - Cornell University : arXiv.org, 2020. - 13 pp..
Programm
When it was introduced in a technical report in May 1994, no one could have foreseen the impact the Burrows-Wheeler Transform (BWT) would have far beyond the field of data compression for which it was originally intended. Of course it first made a significant impact on compression, both in theory and in practice (e.g., as the basis for bzip2). New horizons opened up in 2000 with the introduction of the FM-index, a compressed suffix array based on the BWT. Among other applications, in the next decade FM-indexes became the heart of the DNA aligners such as Bowtie, BWA and SOAP 2 that helped pave the way for the genomics revolution.
Generalizations of BWT to labelled trees, de Bruijn graphs, automata, haplotype sequences and genomic reference graphs have kept the exchange of ideas lively between researchers in algorithms and data structures, bioinformatics, combinatorics on words and information retrieval. Burrows and Wheeler's original technical report is still cited hundreds of times every year, subsequent papers are cited thousands of times, and new results about or using the BWT appear in the top conferences and journals.
By now, 25 years since its publication, probably no one person knows all the results that have been proven about the BWT, but we hope the expertise gathered together at this Dagstuhl Seminar will make progress on the following topics, among others:
FM-indexes for genomic databases: FM-indexes shine for indexing one or a few genomes, but they have not scaled well to indexing the genomic databases that have resulted from high-throughput sequencing technologies. An important problem has been that the suffix array samples used to locate occurrences of patterns must be fairly large or locating becomes very slow. Very recently, a way was discovered to greatly compress also the suffix array sample for repetitive texts, opening the door to indexing thousands of genomes. We expect this seminar will lead to a fuller understanding of this advance and how it can be applied in practice. Another challenge has been beating run-length compression of genomic databases’ BWTs, by identifying additional structure.
More generalizations of the BWT: Many BWT researchers have heard of the generalizations to trees and graphs mentioned above, but it seems few except specialists in algorithms and data structures know about its recent extension to indexed parameterized and order-preserving pattern matching, few except specialists in combinatorics on words know about the alternating BWT, and few except bioinformaticians know about the positional BWT — but each of these may have applications in the other areas. Also, a partially unifying framework has recently been proposed, but there are still many open problems.
New challenges in bioinformatics: Papers on the BWT are published in many venues and no single conference brings together all the experts from algorithms and data structures, combinatorics on words, and theoretical and applied bioinformatics. This disconnect between the areas hurts us all because it prevents knowledge being shared efficiently. The BWT has recently been applied to some surprising bioinformatics problems, such as building ancestral recombination graphs and optical read mapping, and we expect other possibilities will emerge from interdisciplinary discussions.
 Travis Gagie, Giovanni Manzini, Gonzalo Navarro, and Jens Stoye
Travis Gagie, Giovanni Manzini, Gonzalo Navarro, and Jens Stoye
Dagstuhl Seminar 19241 marked the 25th anniversary of the publication of the Burrows-Wheeler Transform (BWT), which has had a huge impact on the fields of data compression, combinatorics on words, compact data structures, and bioinformatics. The 10th anniversary in 2004 was marked by a workshop at the DIMACS Center at Rutgers (http://archive.dimacs.rutgers.edu/Workshops/BWT ) organized by Paolo Ferragina, Giovanni and S. Muthukrishnan, and it is exciting to see how far we have come. In the past 15 years, interest in the BWT has shifted from data compression to compact data structures and bioinformatics, particularly indexing for DNA read alignment, but seven of the 33 participants of that workshop (including Giovanni) also attended this seminar. Unfortunately, Professor Gørtz fell ill at the last minute and emailed us on June 11th to say she couldn't attend, but everyone else on the final list of invitees was present for at least some of the seminar (although not everyone made it into the photo). In total there were 45 people (listed at the end of this report) from 13 countries, including ten women, six junior researchers and two researchers from industry. By happy coincidence, the seminar started the day after Gonzalo's 50th birthday, so we were able to celebrate that as well. We thank Professor Sadakane for the photos shown in Figures 1 and 2.


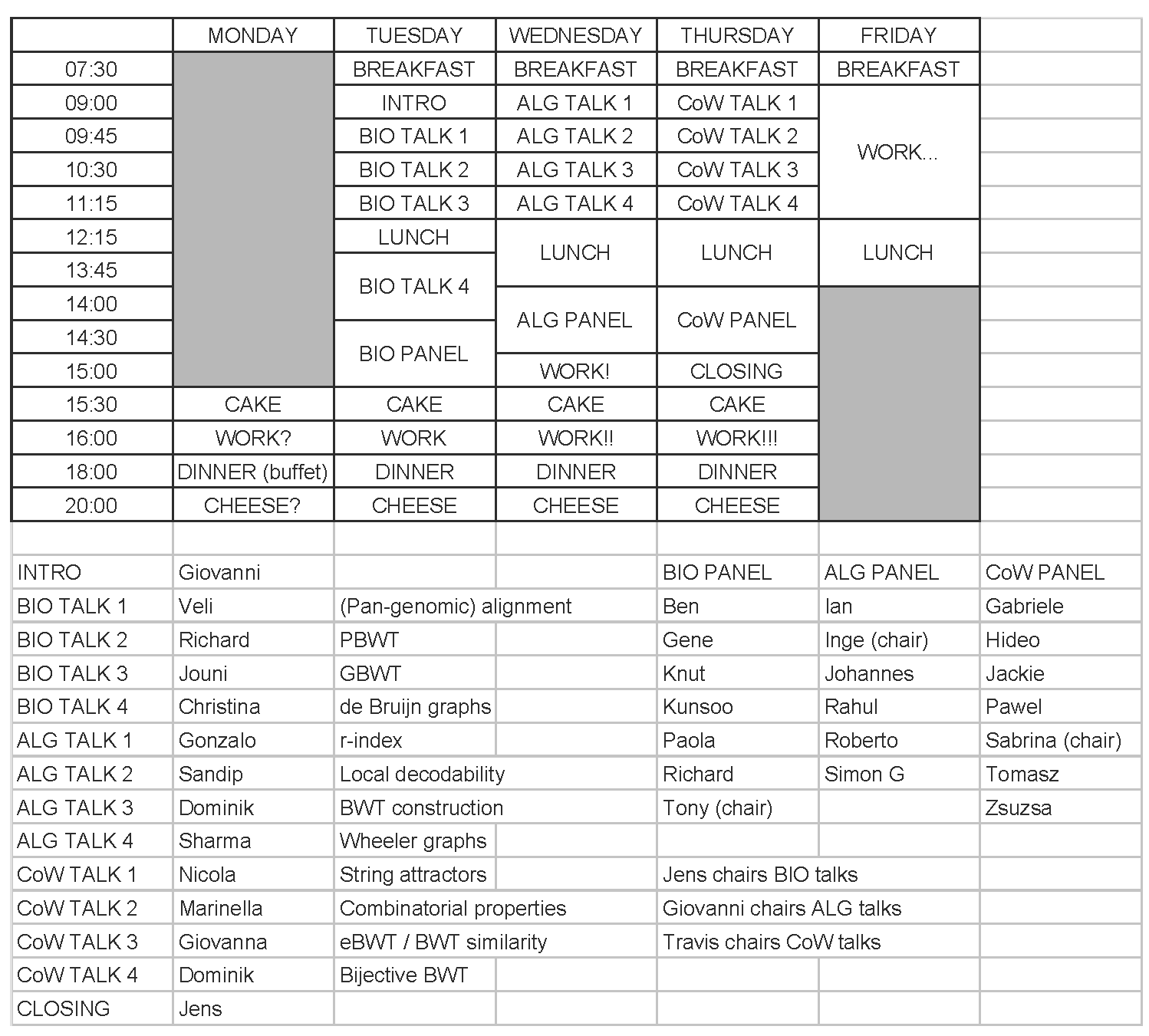
The schedule, shown in Figure 3, featured an introduction, 12 talks, three panel sessions and a closing. The talks were all timely and reflected the active and dynamic research being carried out on the BWT:
- Giovanni's introduction was a more in-depth version of his invited talk from DCC '19;
- Veli Mäkinen surveyed pan-genomic indexing, including work published in BMC Genomics last year;
- Richard Durbin surveyed results based on the positional BWT, published in Bioinformatics in 2014;
- Jouni Sirén presented work included in a Nature Biotechnology article last year;
- Christina Boucher surveyed compact data structures for de Bruijn graphs, including work from an ISMB/ECCB 2019 paper;
- Gonzalo Navarro reviewed BWT-based indexes, including work from a SODA '18 paper;
- Sandip Sinha presented work from a STOC '19 paper;
- Dominik Kempa presented work from another STOC '19 paper;
- Sharma Thankachan presented work from an ESA '19 paper;
- Nicola Prezza presented work from a STOC '18 paper;
- Marinella Sciortino gave a version of her invited lecture for IWOCA '19 a month later;
- Giovanna Rosone presented results about two extensions of the BWT, including work from a WABI '18 paper, now published in Algorithms for Molecular Biology;
- Dominik Klöppl presented work from a CPM '19 paper.
We later received all the abstracts but one.
 Travis Gagie, Giovanni Manzini, Gonzalo Navarro, and Jens Stoye
Travis Gagie, Giovanni Manzini, Gonzalo Navarro, and Jens Stoye
- Jarno Alanko (University of Helsinki, FI) [dblp]
- Hideo Bannai (Kyushu University - Fukuoka, JP) [dblp]
- Paola Bonizzoni (University of Milan-Bicocca, IT) [dblp]
- Christina Boucher (University of Florida - Gainesville, US) [dblp]
- Marilia Braga (Universität Bielefeld, DE) [dblp]
- Anthony J. Cox (Illumina - Saffron Walden, GB) [dblp]
- Fabio Cunial (MPI - Dresden, DE) [dblp]
- Jackie Daykin (Aberystwyth University, GB) [dblp]
- Richard Durbin (University of Cambridge, GB) [dblp]
- Gabriele Fici (University of Palermo, IT) [dblp]
- Johannes Fischer (TU Dortmund, DE) [dblp]
- Travis Gagie (Universidad Diego Portales, CL) [dblp]
- Pawel Gawrychowski (University of Wroclaw, PL) [dblp]
- Simon Gog (eBay Inc - San Jose, US) [dblp]
- Roberto Grossi (University of Pisa, IT) [dblp]
- Wing-Kai Hon (National Tsing Hua University - Hsinchu, TW) [dblp]
- Tomohiro I (Kyushu Institute of Technology - Fukuoka, JP) [dblp]
- Juha Kärkkäinen (University of Helsinki, FI) [dblp]
- Dominik Kempa (University of Warwick - Coventry, GB) [dblp]
- Tomasz Kociumaka (Bar-Ilan University - Ramat Gan, IL) [dblp]
- Dominik Köppl (Kyushu University - Fukuoka, JP) [dblp]
- Ben Langmead (Johns Hopkins University - Baltimore, US) [dblp]
- Zsuzsanna Liptak (University of Verona, IT) [dblp]
- Veli Mäkinen (University of Helsinki, FI) [dblp]
- Sabrina Mantaci (University of Palermo, IT) [dblp]
- Giovanni Manzini (University of Eastern Piedmont - Alessandria, IT) [dblp]
- Ian Munro (University of Waterloo, CA) [dblp]
- Gene Myers (MPI - Dresden, DE) [dblp]
- Gonzalo Navarro (University of Chile - Santiago de Chile, CL) [dblp]
- Yakov Nekrich (University of Waterloo, CA) [dblp]
- Enno Ohlebusch (Universität Ulm, DE) [dblp]
- Kunsoo Park (Seoul National University, KR) [dblp]
- Nicola Prezza (University of Pisa, IT) [dblp]
- Knut Reinert (FU Berlin, DE) [dblp]
- Giovanna Rosone (University of Pisa, IT) [dblp]
- Kunihiko Sadakane (University of Tokyo, JP) [dblp]
- Leena Salmela (University of Helsinki, FI) [dblp]
- Marinella Sciortino (University of Palermo, IT) [dblp]
- Rahul Shah (Louisiana State University - Baton Rouge, US) [dblp]
- Sandip Sinha (Columbia University - New York, US) [dblp]
- Jouni Sirén (University of California - Santa Cruz, US) [dblp]
- Tatiana Starikovskaya (ENS - Paris, FR) [dblp]
- Jens Stoye (Universität Bielefeld, DE) [dblp]
- Sharma V. Thankachan (University of Central Florida - Orlando, US) [dblp]
- Rossano Venturini (University of Pisa, IT) [dblp]
Klassifikation
- bioinformatics
- data bases / information retrieval
- data structures / algorithms / complexity
Schlagworte
- Burrows-Wheeler Transform
- Data Compression
- Indexing
- Sequence Alignment






 Creative Commons BY 3.0 DE
Creative Commons BY 3.0 DE
